Welcome!
This site is a resource for scientists wishing to use derivatives of the Lactococcus lactis (Ll.ltrB) group II intron for genome editing. As the target specificity of this broad host range insertional element is determined by base-pairing between the intron RNA and DNA target, it can be rationally changed by altering the sequence of the intron encoding RNA on purpose-built plasmids. Several such vectors have been constructed that facilitate this process of which the ClosTron was specifically designed for use in Clostridia and related bacteria [1]. This site is maintained by Nottingham researchers at the Synthetic Biology Research Centre (SBRC) and was established to support ClosTron mutagenesis but is also of computational use to those wishing to redesign the targets of other related Targetron systems.
ClosTron Mutagenesis
Here you will find what you need to design, build and test the ClosTron plasmids needed for mutagenesis, including background information, protocols and a computational, intron design tool.

Modular Plasmids
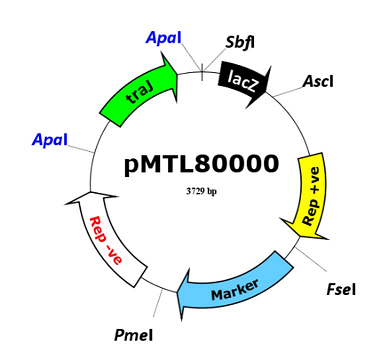
The ClosTron represents an application specific module localised to a pMTL80000 modular clostridia-E. coli shuttle plasmid. Details of the pMTL80000 vectors, together with a series of additional derivative vectors and systems, may be found available at www.plasmidvectors.com.